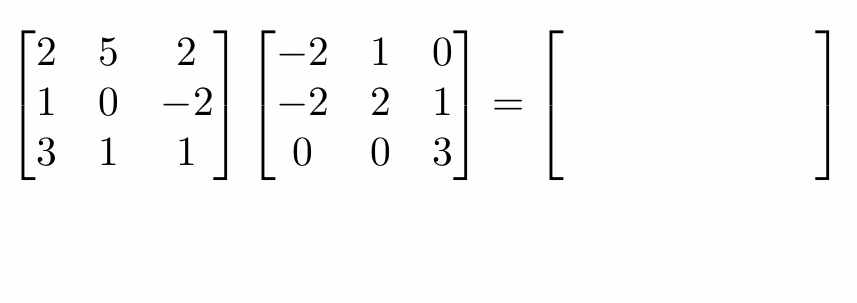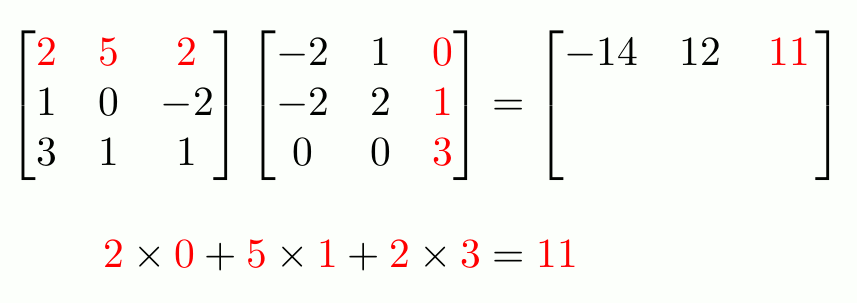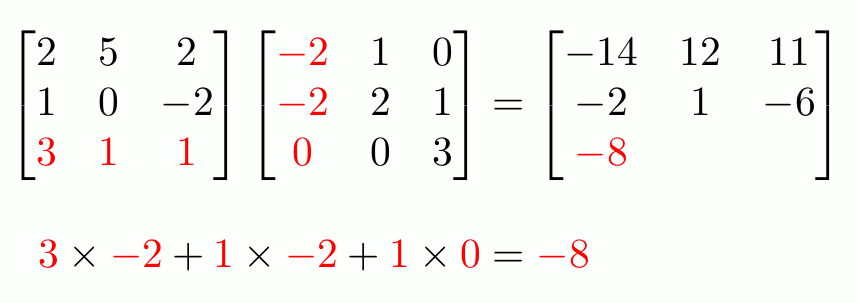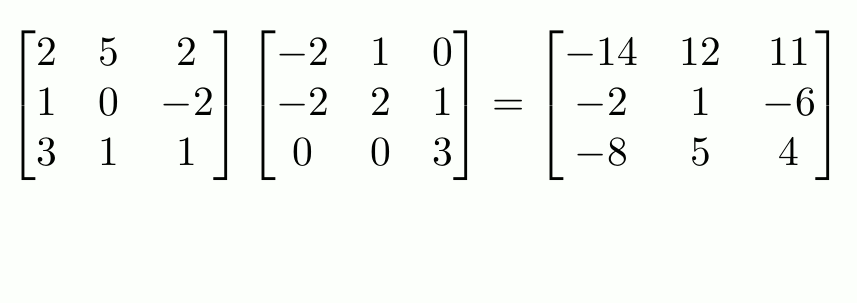Data Structures & Types
CS&SS 508 • Lecture 6
Jess Kunke (slides adapted from Victoria Sass)
Roadmap
Last time, we learned:
- Importing and Exporting Data
- Tidying and Reshaping Data
- Types of Data
- Wrangling Date/Date-Time Data
Today, we will cover:
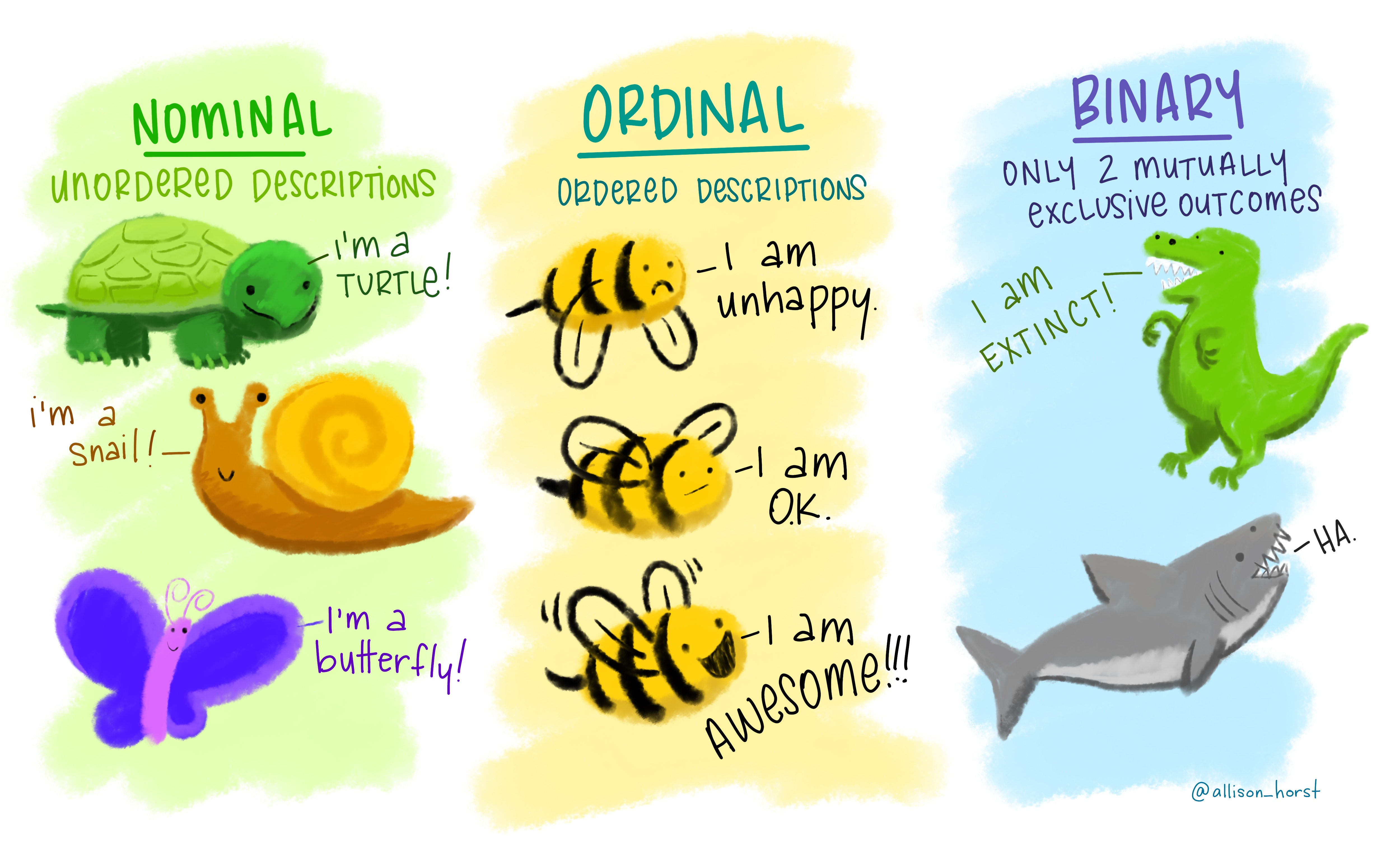
- Types of Data
- Factors
- Numbers
- Missing Values
- Data Structures
- Vectors
- Matrices
- Lists
This week we start getting more into the weeds of programming in R.
These skills will help you understand some of R’s quirks, how to troubleshoot errors when they arise, and how to write more efficient and automated code that does a lot of work for you!
Data types in R
Returning, once again, to our list of data types in R:
- Logicals
- Factors
- Date/Date-time
- Numbers
- Missing Values
- Strings
Data types in R
Returning, once again, to our list of data types in R:
- Logicals
- Factors
- Date/Date-time
- Numbers
- Missing Values
- Strings
Data types in R
Returning, once again, to our list of data types in R:
- Logicals
- Factors
- Date/Date-time
- Numbers
- Missing Values
- Strings

Data types in R
Returning, once again, to our list of data types in R:
Logicals- Factors
Date/Date-time- Numbers
- Missing Values
- Strings
Working with Factors
Why Use Factors?
Factors are a special class of data specifically for categorical variables1 which have a fixed, known, and mutually exclusive set of possible values2.
Imagine we have a variable that records the month that an event occurred.
The two main issues with coding this simply as a character string:
- It doesn’t help catch spelling errors
- Characters are sorted alphabetically, which is not necessarily intuitive or useful for your variable
Factors
Factors have an additional specification called levels. These are the categories of the categorical variable. We can create a vector of the levels first:
And then we can create a factor like so:
> [1] Dec Apr Jan Mar
> Levels: Jan Feb Mar Apr May Jun Jul Aug Sep Oct Nov DecWe can see that the levels specify in which order the categories should be displayed:
Creating Factors
factor is Base R’s function for creating factors while fct is forcats function for making factors. A couple of things to note about their differences:
factor
- Any values not specified as a level will be silently converted to
NA - Without specified levels, they’ll be created from the data in alphabetical order1
fct
- Will send a error message if a value exists outside the specified levels
- Without specified levels, they’ll be created from the data in order of first appearance
You can create a factor by specifying col_factor() when reading in data with readr:
If you need to access the levels directly you can use the Base R function levels().
Changing the Order of Levels
One of the more common data manipulations you’ll want to do with factors is to change the ordering of the levels. This could be to put them in a more intuitive order but also to make a visualization clearer and more impactful.
Let’s use a subset of the General Social Survey1 data to see what this might look like.
> # A tibble: 21,483 × 9
> year marital age race rincome partyid relig denom tvhours
> <int> <fct> <int> <fct> <fct> <fct> <fct> <fct> <int>
> 1 2000 Never married 26 White $8000 to 9999 Ind,near … Prot… Sout… 12
> 2 2000 Divorced 48 White $8000 to 9999 Not str r… Prot… Bapt… NA
> 3 2000 Widowed 67 White Not applicable Independe… Prot… No d… 2
> 4 2000 Never married 39 White Not applicable Ind,near … Orth… Not … 4
> 5 2000 Divorced 25 White Not applicable Not str d… None Not … 1
> 6 2000 Married 25 White $20000 - 24999 Strong de… Prot… Sout… NA
> 7 2000 Never married 36 White $25000 or more Not str r… Chri… Not … 3
> 8 2000 Divorced 44 White $7000 to 7999 Ind,near … Prot… Luth… NA
> 9 2000 Married 44 White $25000 or more Not str d… Prot… Other 0
> 10 2000 Married 47 White $25000 or more Strong re… Prot… Sout… 3
> # ℹ 21,473 more rowsChanging the Order of Levels
There are four related functions to change the level ordering in forcats.
fct_reorder()
- 1
-
factoris the factor to reorder (or a character string to be turned into a factor) - 2
-
ordering_vectorspecifies how to reorderfactor - 3
-
optional_functionis applied if there are multiple values ofordering_vectorfor each value offactor(the default is to take the median)
fct_relevel()
- 4
-
valueis either a function (i.e.sort) or a character level (default is to move it to the front of the vector) - 5
-
placementis an optional vector index where the level should be placed
fct_reorder2()
fct_reorder2(.f = factor,
6 .x = vector1,
.y = vector2)- 6
-
fct_reorder2reordersfactorby the values ofvector2associated with the largest values ofvector1.
fct_infreq()
7fct_infreq(.f = factor)- 7
-
fct_infreqreordersfactorin decreasing frequency. See other variations here. Use withfct_rev()for increasing frequency.
Changing the Order of Levels
There are four related functions to change the level ordering in forcats.
fct_reorder() is for reordering levels by sorting along another variable
Without fct_reorder()
Code
With fct_reorder()
fct_relevel() allows you to reorder the levels by hand
Without fct_relevel()
Code
With fct_relevel()
fct_reorder2() is like fct_reorder(), except when the factor is mapped to a non-position aesthetic such as color
Without fct_reorder2()
Code
With fct_reorder()
fct_infreq() reorders levels by the number of observations within each level (largest first)
Without fct_infreq()
With fct_infreq()
Question for you: what does fct_rev() do here?
Changing the Value of Levels
You may also want to change the actual values of your factor levels. The main way to do this is fct_recode().
8gss_cat |> count(partyid)- 8
-
You can use
count()to get the full list of levels for a variable and their respective counts.
> # A tibble: 10 × 2
> partyid n
> <fct> <int>
> 1 No answer 154
> 2 Don't know 1
> 3 Other party 393
> 4 Strong republican 2314
> 5 Not str republican 3032
> 6 Ind,near rep 1791
> 7 Independent 4119
> 8 Ind,near dem 2499
> 9 Not str democrat 3690
> 10 Strong democrat 3490fct_recode()
gss_cat |>
mutate(
partyid = fct_recode(partyid,
"Republican, strong" = "Strong republican",
"Republican, weak" = "Not str republican",
"Independent, near rep" = "Ind,near rep",
"Independent, near dem" = "Ind,near dem",
"Democrat, weak" = "Not str democrat",
"Democrat, strong" = "Strong democrat"
)
) |>
count(partyid)> # A tibble: 10 × 2
> partyid n
> <fct> <int>
> 1 No answer 154
> 2 Don't know 1
> 3 Other party 393
> 4 Republican, strong 2314
> 5 Republican, weak 3032
> 6 Independent, near rep 1791
> 7 Independent 4119
> 8 Independent, near dem 2499
> 9 Democrat, weak 3690
> 10 Democrat, strong 3490Some features of fct_recode():
- Will leave the levels that aren’t explicitly mentioned, as is.
- Will warn you if you accidentally refer to a level that doesn’t exist.
- You can combine groups by assigning multiple old levels to the same new level.
fct_collapse()
A useful variant of fct_recode() is fct_collapse() which will allow you to collapse a lot of levels at once.
gss_cat |>
mutate(
partyid = fct_collapse(partyid,
"other" = c("No answer", "Don't know", "Other party"),
"rep" = c("Strong republican", "Not str republican"),
"ind" = c("Ind,near rep", "Independent", "Ind,near dem"),
"dem" = c("Not str democrat", "Strong democrat")
)
) |>
count(partyid)> # A tibble: 4 × 2
> partyid n
> <fct> <int>
> 1 other 548
> 2 rep 5346
> 3 ind 8409
> 4 dem 7180fct_lump_*
Sometimes you’ll have several levels of a variable that have a small enough N to warrant grouping them together into an other category. The family of fct_lump_* functions are designed to help with this.
gss_cat |>
9 mutate(relig = fct_lump_n(relig, n = 10)) |>
count(relig, sort = TRUE)- 9
-
Other functions include:
fct_lump_min(),fct_lump_prop(),fct_lump_lowfreq(). Read more about them here.
> # A tibble: 10 × 2
> relig n
> <fct> <int>
> 1 Protestant 10846
> 2 Catholic 5124
> 3 None 3523
> 4 Christian 689
> 5 Other 458
> 6 Jewish 388
> 7 Buddhism 147
> 8 Inter-nondenominational 109
> 9 Moslem/islam 104
> 10 Orthodox-christian 95Ordered Factors
So far we’ve mostly been discussing how to code nominal variables, or categorical variables that have no inherent ordering.
If you want to specify that your factor has a strict order you can classify it as a ordered factor.
10ordered(c("a", "b", "c"))- 10
- Ordered factors imply a strict ordering and equal distance between levels: the first level is “less than” the second level by the same amount that the second level is “less than” the third level, and so on.
> [1] a b c
> Levels: a < b < cIn practice there are only two ways in which ordered factors are different than factors:
scale_color_viridis()/scale_fill_viridis()will be used automatically when mapping an ordered factored inggplot2because it implies an ordered ranking- If you use an ordered function in a linear model, it will use “polygonal contrasts”. You can learn more about what this means here.
Numbers
Numbers, Two Ways
R has two types of numeric variables: double and integer.
- Integers must be round numbers while doubles can be decimals
- They are stored differently, and arithmetic works a bit differently with integers vs doubles
- Continuous values must be stored as doubles
- Discrete values can be integers, doubles, or factors

Numbers Coded as Character Strings
Oftentimes numerical data is coded as a string so you’ll need to use the appropriate parsing function to read it in in the correct form.
If you have values with extraneous non-numerical text you want to ignore there’s a separate function for that.
count()
A very useful and common exploratory data analysis tool is to check the relative sums of different categories of a variable. That’s what count() is for!
library(nycflights13)
data(flights)
1flights |> count(origin)- 1
-
Add the argument
sort = TRUEto see the most common values first (i.e. arranged in descending order). . . .
> # A tibble: 3 × 2
> origin n
> <chr> <int>
> 1 EWR 120835
> 2 JFK 111279
> 3 LGA 104662This is functionally the same as grouping and summarizing with n().
- 2
-
n()is a special summary function that doesn’t take any arguments and instead accesses information about the “current” group. This means that it only works inside dplyr verbs. - 3
- You can do this longer version if you also want to compute other summaries simultaneously.
> # A tibble: 3 × 2
> origin n
> <chr> <int>
> 1 EWR 120835
> 2 LGA 104662
> 3 JFK 111279n_distinct()
Use this function if you want the count the number of distinct (unique) values of one or more variables.
Say we’re interested in which destinations are served by the most carriers:
Weighted Counts
A weighted count is simply a grouped sum, therefore count has a wt argument to allow for the shorthand.
How many miles did each plane fly?
> # A tibble: 4,044 × 2
> tailnum miles
> <chr> <dbl>
> 1 N14228 171713
> 2 N24211 172934
> 3 N619AA 32141
> 4 N804JB 311992
> 5 N668DN 50352
> 6 N39463 169905
> 7 N516JB 359585
> 8 N829AS 52549
> 9 N593JB 377619
> 10 N3ALAA 67925
> # ℹ 4,034 more rowsThis is equivalent to:
Other Useful Arithmetic Functions
In addition to the standards (+, -, /, *, ^), R has many other useful arithmetic functions.
Pairwise min/max
- 6
-
pmin()returns the smallest value in each row.min(), by contrast, finds the smallest observation given a number of rows. - 7
-
pmax()returns the largest value in each row.max(), by contrast, finds the largest observation given a number of rows.
> # A tibble: 3 × 4
> x y min max
> <dbl> <dbl> <dbl> <dbl>
> 1 1 3 1 3
> 2 5 2 2 5
> 3 7 NA 7 7Other Useful Arithmetic Functions
Modular arithmetic
We can see how this can be useful in our flights data which has curiously stored time:
> # A tibble: 336,776 × 3
> sched_dep_time hour minute
> <int> <dbl> <dbl>
> 1 515 5 15
> 2 529 5 29
> 3 540 5 40
> 4 545 5 45
> 5 600 6 0
> 6 558 5 58
> 7 600 6 0
> 8 600 6 0
> 9 600 6 0
> 10 600 6 0
> # ℹ 336,766 more rowsOther Useful Arithmetic Functions
Logarithms1
11log2(c(2, 4, 8))- 11
-
Easy to interpret because a difference of 1 on the log scale corresponds to doubling on the original scale and a difference of -1 corresponds to halving. Inverse is
2^.
> [1] 1 2 312log10(c(10, 100, 1000))- 12
-
Easy to back-transform because everything is on the order of 10. Inverse is
10^.
> [1] 1 2 3Other Useful Arithmetic Functions
Cumulative and Rolling Aggregates
Base R provides cumsum(), cumprod(), cummin(), cummax() for running, or cumulative, sums, products, mins and maxes. dplyr provides cummean() for cumulative means.
13cumsum(1:15)- 13
-
cumsum()is the most common in practice.
> [1] 1 3 6 10 15 21 28 36 45 55 66 78 91 105 120For complex rolling/sliding aggregates, check out the slidr package.
Other Useful Arithmetic Functions
Numeric Ranges
x <- c(1, 2, 5, 10, 15, 20)
14cut(x, breaks = c(0, 5, 10, 15, 20))- 14
-
cut()breaks up (aka bins) a numeric vector into discrete buckets
> [1] (0,5] (0,5] (0,5] (5,10] (10,15] (15,20]
> Levels: (0,5] (5,10] (10,15] (15,20]15cut(x, breaks = c(0, 5, 10, 100))- 15
- The bins don’t have to be the same size.
> [1] (0,5] (0,5] (0,5] (5,10] (10,100] (10,100]
> Levels: (0,5] (5,10] (10,100]cut(x,
breaks = c(0, 5, 10, 15, 20),
16 labels = c("sm", "md", "lg", "xl")
)- 16
- You can optionally supply your own labels. Note that there should be one less labels than breaks.
> [1] sm sm sm md lg xl
> Levels: sm md lg xl17y <- c(NA, -10, 5, 10, 30)
cut(y, breaks = c(0, 5, 10, 15, 20))- 17
-
Any values outside of the range of the breaks will become
NA.
> [1] <NA> <NA> (0,5] (5,10] <NA>
> Levels: (0,5] (5,10] (10,15] (15,20]Rounding
round() allows us to round to a certain decimal place. Without specifying an argument for the digits argument it will round to the nearest integer.
Rounding
What’s going on here?
round() uses what’s known as “round half to even” or Banker’s rounding: if a number is half way between two integers, it will be rounded to the even integer. This is a good strategy because it keeps the rounding unbiased: half of all 0.5s are rounded up, and half are rounded down.
Summary Functions
Central Tendency
20x <- sample(1:500, size = 100, replace = TRUE)
mean(x)- 20
-
sample()takes a vector of data, and samplessizeelements from it, with replacement ifreplaceequalsTRUE.
> [1] 253.4821quantile(x, .95)- 21
-
A generalization of the median:
quantile(x, 0.95)will find the value that’s greater than 95% of the values;quantile(x, 0.5)is equivalent to the median.
> 95%
> 490Summary Functions
Measures of Spread/Variation
22IQR(x)- 22
-
Equivalent to
quantile(x, 0.75) - quantile(x, 0.25)and gives you the range that contains the middle 50% of the data.
> [1] 263.5Common Numerical Manipulations
These formulas can be used in a summary call but are also useful with mutate(), particularly if being applied to grouped data.
Summary Functions
Positions
These are all really helpful but is there a good summary descriptive statistics function?
Basic summary statistics
> Sepal.Length Sepal.Width Petal.Length Petal.Width
> Min. :4.300 Min. :2.000 Min. :1.000 Min. :0.100
> 1st Qu.:5.100 1st Qu.:2.800 1st Qu.:1.600 1st Qu.:0.300
> Median :5.800 Median :3.000 Median :4.350 Median :1.300
> Mean :5.843 Mean :3.057 Mean :3.758 Mean :1.199
> 3rd Qu.:6.400 3rd Qu.:3.300 3rd Qu.:5.100 3rd Qu.:1.800
> Max. :7.900 Max. :4.400 Max. :6.900 Max. :2.500
> Species
> setosa :50
> versicolor:50
> virginica :50
>
>
> Better summary statistics
A basic example:
| Name | iris |
| Number of rows | 150 |
| Number of columns | 5 |
| _______________________ | |
| Column type frequency: | |
| factor | 1 |
| numeric | 4 |
| ________________________ | |
| Group variables | None |
Variable type: factor
| skim_variable | n_missing | complete_rate | ordered | n_unique | top_counts |
|---|---|---|---|---|---|
| Species | 0 | 1 | FALSE | 3 | set: 50, ver: 50, vir: 50 |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| Sepal.Length | 0 | 1 | 5.84 | 0.83 | 4.3 | 5.1 | 5.80 | 6.4 | 7.9 | ▆▇▇▅▂ |
| Sepal.Width | 0 | 1 | 3.06 | 0.44 | 2.0 | 2.8 | 3.00 | 3.3 | 4.4 | ▁▆▇▂▁ |
| Petal.Length | 0 | 1 | 3.76 | 1.77 | 1.0 | 1.6 | 4.35 | 5.1 | 6.9 | ▇▁▆▇▂ |
| Petal.Width | 0 | 1 | 1.20 | 0.76 | 0.1 | 0.3 | 1.30 | 1.8 | 2.5 | ▇▁▇▅▃ |
Better summary statistics
A more complex example:
| Name | starwars |
| Number of rows | 87 |
| Number of columns | 14 |
| _______________________ | |
| Column type frequency: | |
| character | 8 |
| list | 3 |
| numeric | 3 |
| ________________________ | |
| Group variables | None |
Variable type: character
| skim_variable | n_missing | complete_rate | min | max | empty | n_unique | whitespace |
|---|---|---|---|---|---|---|---|
| name | 0 | 1.00 | 3 | 21 | 0 | 87 | 0 |
| hair_color | 5 | 0.94 | 4 | 13 | 0 | 11 | 0 |
| skin_color | 0 | 1.00 | 3 | 19 | 0 | 31 | 0 |
| eye_color | 0 | 1.00 | 3 | 13 | 0 | 15 | 0 |
| sex | 4 | 0.95 | 4 | 14 | 0 | 4 | 0 |
| gender | 4 | 0.95 | 8 | 9 | 0 | 2 | 0 |
| homeworld | 10 | 0.89 | 4 | 14 | 0 | 48 | 0 |
| species | 4 | 0.95 | 3 | 14 | 0 | 37 | 0 |
Variable type: list
| skim_variable | n_missing | complete_rate | n_unique | min_length | max_length |
|---|---|---|---|---|---|
| films | 0 | 1 | 24 | 1 | 7 |
| vehicles | 0 | 1 | 11 | 0 | 2 |
| starships | 0 | 1 | 16 | 0 | 5 |
Variable type: numeric
| skim_variable | n_missing | complete_rate | mean | sd | p0 | p25 | p50 | p75 | p100 | hist |
|---|---|---|---|---|---|---|---|---|---|---|
| height | 6 | 0.93 | 174.60 | 34.77 | 66 | 167.0 | 180 | 191.0 | 264 | ▂▁▇▅▁ |
| mass | 28 | 0.68 | 97.31 | 169.46 | 15 | 55.6 | 79 | 84.5 | 1358 | ▇▁▁▁▁ |
| birth_year | 44 | 0.49 | 87.57 | 154.69 | 8 | 35.0 | 52 | 72.0 | 896 | ▇▁▁▁▁ |
skim function
Highlights of this summary statistics function:
- provides a larger set of statistics than
summary()including number missing, complete, n, sd, histogram for numeric data - presentation is in a compact, organized format
- reports each data type separately
- handles a wide range of data classes including dates, logicals, strings, lists and more
- can be used with
summary()for an overall summary of the data (w/o specifics about columns) - individual columns can be selected for a summary of only a subset of the data
- handles grouped data
- behaves nicely in pipelines
- produces knitted results for documents
- easily and highly customizable (i.e. specify your own statistics and classes)
Missing Values
Explicit Missing Values
An explicit missing value is the presence of an absence.
In other words, an explicit missing value is one in which you see an NA.
Depending on the reason for its missingness, there are different ways to deal with NAs.
Data Entry Shorthand
If your data were entered by hand and NAs merely represent a value being carried forward from the last entry then you can use fill() to help complete your data.
treatment |>
1 fill(everything())- 1
-
fill()takes one or more variables (in this caseeverything(), which means all variables), and by default fills them in downwards. If you have a different issue you can change the.directionargument to"up","downup", or"updown".
> # A tibble: 4 × 3
> person treatment response
> <chr> <dbl> <dbl>
> 1 Derrick Whitmore 1 7
> 2 Derrick Whitmore 2 10
> 3 Katherine Burke 3 10
> 4 Katherine Burke 1 4Explicit Missing Values
Represent A Fixed Value
Other times an NA represents some fixed value, usually 0.
x <- c(1, 4, 5, 7, NA)
2coalesce(x, 0)- 2
-
coalesce()in thedplyrpackage takes a vector as the first argument and will replace any missing values with the value provided in the second argument.
> [1] 1 4 5 7 0Explicit Missing Values
NaNs
A special sub-type of missing value is an NaN, or Not a Number.
These generally behave similar to NAs and are likely the result of a mathematical operation that has an indeterminate result:
If you need to explicitly identify an NaN you can use is.nan().
Implicit NAs
An implicit missing value is the absence of a presence.
We’ve seen a couple of ways that implicit NAs can be made explicit in previous lectures: pivoting and joining.
For example, if we really look at the dataset below, we can see that there are missing values that don’t appear as NA merely due to the current structure of the data.
Implicit NAs
tidyr::complete() allows you to generate explicit missing values by providing a set of variables that define the combination of rows that should exist.
Missing Factor Levels
The last type of missingness is a theoretical level of a factor that doesn’t have any observations.
For instance, we have this health dataset and we’re interested in smokers:
3levels(health$smoker)- 3
- This dataset only contains non-smokers, but we know that smokers exist; the group of smokers is simply empty.
> [1] "yes" "no"
4health |> count(smoker, .drop = FALSE)- 4
-
We can request
count()to keep all the groups, even those not seen in the data by using.drop = FALSE.
> # A tibble: 2 × 2
> smoker n
> <fct> <int>
> 1 yes 0
> 2 no 5Missing Factors in Plots
This sample principle applies when visualizing a factor variable, which will automatically drop levels that don’t have any values. Use drop_values = FALSE in the appropriate scale to display implicit NAs.
Testing Data Types
There are also functions to test for certain data types:
Going deeper into the abyss (aka NAs)

Going deeper into the abyss (aka NAs)
A lot has been written about NAs and if they are a feature of your data you’re likely going to have to spend a great deal of time thinking about how they arose1 and if/how they bias your data.
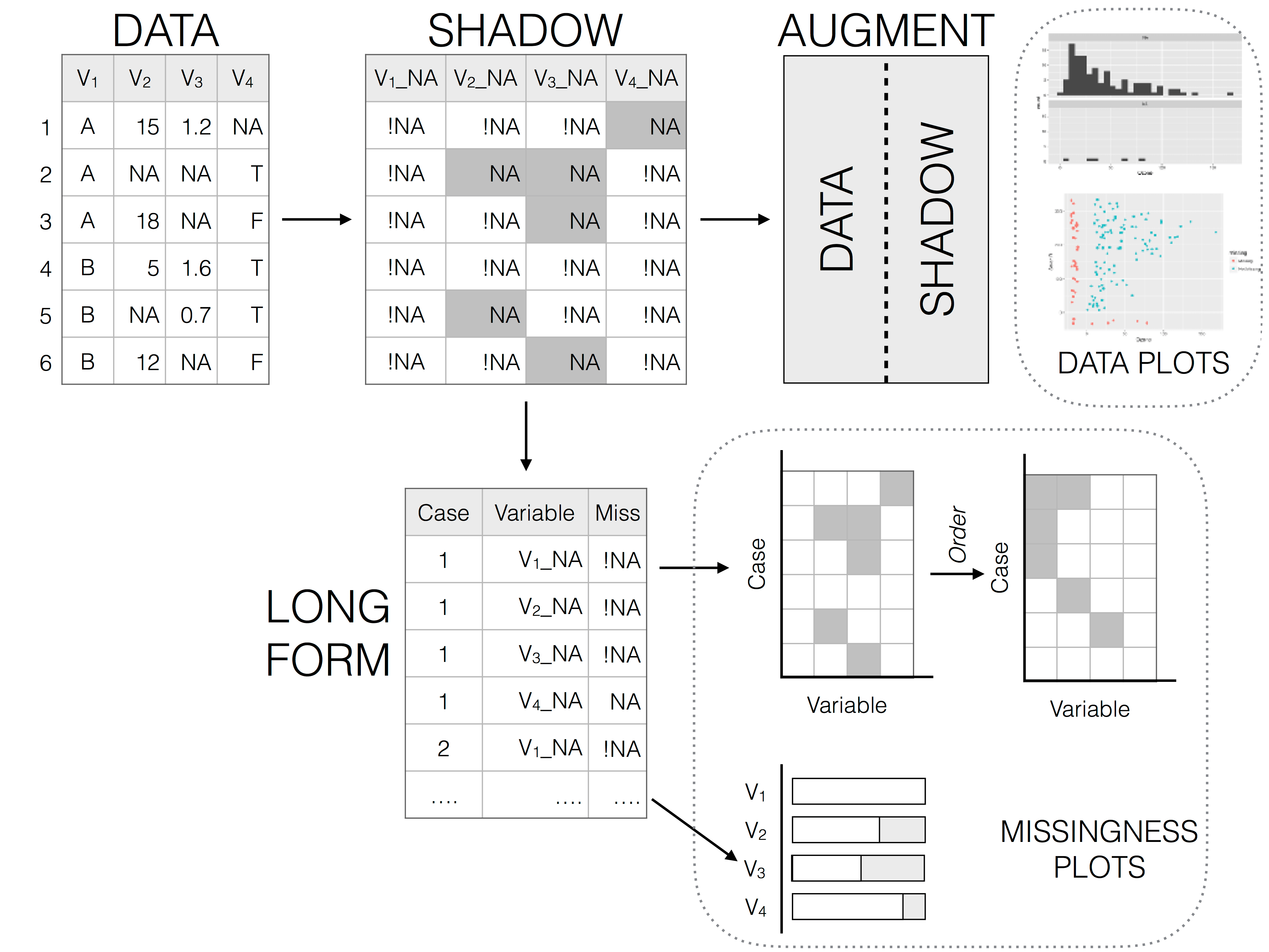
The best package for really exploring your NAs is naniar, which provides tidyverse-style syntax for summarizing, visualizing, and manipulating missing data.
It provides the following for missing data:
- a special data structure
- shorthand and numerical summaries (in variables and cases)
- visualizations

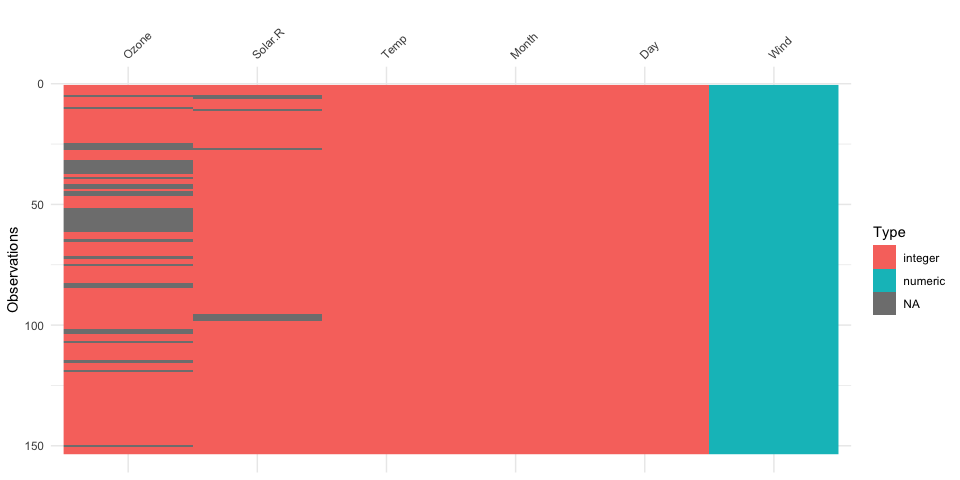
naniar examples
visdat example
Break!
Vectors
Making Vectors
In R, we call a set of values of the same type a vector. We can create vectors using the c() function (“c” for combine or concatenate).
All elements of a vector are the same type (e.g. numeric or character)!
Character data is the lowest denomination so anything mixed with it will be converted to a character.
Generating Numeric Vectors
There are shortcuts for generating numeric vectors:
1seq(-3, 6, by = 1.75)- 1
- Sequence from -3 to 6, increments of 1.75
> [1] -3.00 -1.25 0.50 2.25 4.00 5.75You can also assign values to a vector using Base R indexing rules.
Element-wise Vector Math
When doing arithmetic operations on vectors, R handles these element-wise:
Other common operations: *, /, exp() = \(e^x\), log() = \(\log_e(x)\)
Recycling Rules
R handles mismatched lengths of vectors by recycling, or repeating, the short vector.
You generally only want to recycle scalars, or vectors of length 1. Technically, however, R will recycle any vector that’s shorter in length (and it won’t always give you a warning that that’s what it’s doing, i.e. if the longer vector is not a multiple of the shorter vector).
Recycling with Logicals
The same rules apply to logical operations which can lead to unexpected results without warning.
For example, take this code which attempts to find all flights in January and February:
flights |>
mutate(rowID = 1:nrow(flights)) |>
relocate(rowID) |>
7 filter(month == c(1, 2))- 7
-
A common mistake is to mix up
==with%in%. This code will actually find flights in odd numbered rows that departed in January and flights in even numbered rows that departed in February. Unfortunately there’s no warning becauseflightshas an even number of rows.
> # A tibble: 25,977 × 20
> rowID year month day dep_time sched_dep_time dep_delay arr_time
> <int> <int> <int> <int> <int> <int> <dbl> <int>
> 1 1 2013 1 1 517 515 2 830
> 2 3 2013 1 1 542 540 2 923
> 3 5 2013 1 1 554 600 -6 812
> 4 7 2013 1 1 555 600 -5 913
> 5 9 2013 1 1 557 600 -3 838
> 6 11 2013 1 1 558 600 -2 849
> 7 13 2013 1 1 558 600 -2 924
> 8 15 2013 1 1 559 600 -1 941
> 9 17 2013 1 1 559 600 -1 854
> 10 19 2013 1 1 600 600 0 837
> # ℹ 25,967 more rows
> # ℹ 12 more variables: sched_arr_time <int>, arr_delay <dbl>, carrier <chr>,
> # flight <int>, tailnum <chr>, origin <chr>, dest <chr>, air_time <dbl>,
> # distance <dbl>, hour <dbl>, minute <dbl>, time_hour <dttm>To protect you from this type of silent failure, most tidyverse functions use a stricter form of recycling that only recycles single values. However, when using base R functions like ==, this protection is not built in.
Example: Standardizing Data
Let’s say we had some test scores and we wanted to put these on a standardized scale:
\[z_i = \frac{x_i - \text{mean}(x)}{\text{SD}(x)}\]
Math with Missing Values
Even one NA “poisons the well”: You’ll get NA out of your calculations unless you add the extra argument na.rm = TRUE (available in some functions):
Subsetting Vectors
Recall, we can subset a vector in a number of ways:
- Passing a single index or vector of entries to keep:
> [1] "Andre" "Brady"- Passing a single index or vector of entries to drop:
- Passing a logical condition:
8first_names[nchar(first_names) == 7]- 8
-
nchar()counts the number of characters in a character string.
> [1] "Cecilia" "Francie"Matrices
Matrices: Two Dimensions
Matrices extend vectors to two dimensions: rows and columns. We can construct them directly using matrix().
R fills in a matrix column-by-column (not row-by-row!)
Similar to vectors, you can make assignments using Base R indexing methods.
Binding Vectors
We can also make matrices by binding vectors together with rbind() (row bind) and cbind() (column bind).
Subsetting Matrices
We subset matrices using the same methods as with vectors, except we index them with [rows, columns]1:
Matrices Becoming Vectors
If a matrix ends up having just one row or column after subsetting, by default R will make it into a vector.
Matrix Data Type Warning
Matrices can contain numeric, integer, factor, character, or logical. But just like vectors, all elements must be the same data type.
In this case, everything was converted to characters!
Matrix Dimension Names
We can access dimension names or name them ourselves:
> Number Name
> First "1" "apple"
> Last "2" "banana"11bad_matrix[ ,"Name", drop = FALSE]- 11
-
drop = FALSEmaintains the matrix structure; whendrop = TRUE(the default) it will be converted to a vector.
> Name
> First "apple"
> Last "banana"Matrix Arithmetic
Matrices of the same dimensions can have math performed element-wise with the usual arithmetic operators:
“Proper” Matrix Math
To do matrix transpositions, use t().
“Proper” Matrix Math
To invert an invertible square matrix1, use solve().
Matrices vs. Data.frames and Tibbles
All of these structures display data in two dimensions
matrix- Base
R - Single data type allowed
- Base
data.frame- Base
R - Stores multiple data types
- Default for data storage
- Base
tibblestidyverse- Stores multiple data types
- Displays nicely
In practice, data.frames and tibbles are very similar!
Creating data.frames or tibbles
We can create a data.frame or tibble by specifying the columns separately, as individual vectors:
Note: data.frames and tibbles allow for mixed data types!
This distinction leads us to the final data type, of which data.frames and tibbles are a particular subset.
Lists
What are Lists?
Lists are objects that can store multiple types of data.
my_list <- list(first_thing = 1:5,
second_thing = matrix(8:11, nrow = 2),
third_thing = fct(c("apple", "pear", "banana", "apple", "apple")))
my_list> $first_thing
> [1] 1 2 3 4 5
>
> $second_thing
> [,1] [,2]
> [1,] 8 10
> [2,] 9 11
>
> $third_thing
> [1] apple pear banana apple apple
> Levels: apple pear bananaAccessing List Elements
You can access a list element by its name or number in [[ ]], or a $ followed by its name:
Why Two Brackets [[ ]]?
Double brackets get the actual element — as whatever data type it is stored as, in that location in the list.
names() and List Elements
You can use names() to get a vector of list element names:
pluck()
An alternative to using Base R’s [[ ]] is using pluck() from the tidyverse’s purrr package.
obj1 <- list("a", list(1, elt = "foo"))
obj2 <- list("b", list(2, elt = "bar"))
x <- list(obj1, obj2)
x> [[1]]
> [[1]][[1]]
> [1] "a"
>
> [[1]][[2]]
> [[1]][[2]][[1]]
> [1] 1
>
> [[1]][[2]]$elt
> [1] "foo"
>
>
>
> [[2]]
> [[2]][[1]]
> [1] "b"
>
> [[2]][[2]]
> [[2]][[2]][[1]]
> [1] 2
>
> [[2]][[2]]$elt
> [1] "bar"pluck()
An alternative to using Base R’s [[ ]] is using pluck() from the tidyverse’s purrr package.
obj1 <- list("a", list(1, elt = "foo"))
obj2 <- list("b", list(2, elt = "bar"))
x <- list(obj1, obj2)
x> [[1]]
> [[1]][[1]]
> [1] "a"
>
> [[1]][[2]]
> [[1]][[2]][[1]]
> [1] 1
>
> [[1]][[2]]$elt
> [1] "foo"
>
>
>
> [[2]]
> [[2]][[1]]
> [1] "b"
>
> [[2]][[2]]
> [[2]][[2]][[1]]
> [1] 2
>
> [[2]][[2]]$elt
> [1] "bar"pluck()
An alternative to using Base R’s [[ ]] is using pluck() from the tidyverse’s purrr package.
obj1 <- list("a", list(1, elt = "foo"))
obj2 <- list("b", list(2, elt = "bar"))
x <- list(obj1, obj2)
x> [[1]]
> [[1]][[1]]
> [1] "a"
>
> [[1]][[2]]
> [[1]][[2]][[1]]
> [1] 1
>
> [[1]][[2]]$elt
> [1] "foo"
>
>
>
> [[2]]
> [[2]][[1]]
> [1] "b"
>
> [[2]][[2]]
> [[2]][[2]][[1]]
> [1] 2
>
> [[2]][[2]]$elt
> [1] "bar"Example: Regression Output
When you perform linear regression in R, the output is a list!
What does a list object look like?
> List of 12
> $ coefficients : Named num [1:2] 8.284 0.166
> ..- attr(*, "names")= chr [1:2] "(Intercept)" "dist"
> $ residuals : Named num [1:50] -4.62 -5.94 -1.95 -4.93 -2.93 ...
> ..- attr(*, "names")= chr [1:50] "1" "2" "3" "4" ...
> $ effects : Named num [1:50] -108.894 29.866 -0.501 -3.945 -1.797 ...
> ..- attr(*, "names")= chr [1:50] "(Intercept)" "dist" "" "" ...
> $ rank : int 2
> $ fitted.values: Named num [1:50] 8.62 9.94 8.95 11.93 10.93 ...
> ..- attr(*, "names")= chr [1:50] "1" "2" "3" "4" ...
> $ assign : int [1:2] 0 1
> $ qr :List of 5
> ..$ qr : num [1:50, 1:2] -7.071 0.141 0.141 0.141 0.141 ...
> .. ..- attr(*, "dimnames")=List of 2
> .. .. ..$ : chr [1:50] "1" "2" "3" "4" ...
> .. .. ..$ : chr [1:2] "(Intercept)" "dist"
> .. ..- attr(*, "assign")= int [1:2] 0 1
> ..$ qraux: num [1:2] 1.14 1.15
> ..$ pivot: int [1:2] 1 2
> ..$ tol : num 1e-07
> ..$ rank : int 2
> ..- attr(*, "class")= chr "qr"
> $ df.residual : int 48
> $ xlevels : Named list()
> $ call : language lm(formula = speed ~ dist, data = cars)
> $ terms :Classes 'terms', 'formula' language speed ~ dist
> .. ..- attr(*, "variables")= language list(speed, dist)
> .. ..- attr(*, "factors")= int [1:2, 1] 0 1
> .. .. ..- attr(*, "dimnames")=List of 2
> .. .. .. ..$ : chr [1:2] "speed" "dist"
> .. .. .. ..$ : chr "dist"
> .. ..- attr(*, "term.labels")= chr "dist"
> .. ..- attr(*, "order")= int 1
> .. ..- attr(*, "intercept")= int 1
> .. ..- attr(*, "response")= int 1
> .. ..- attr(*, ".Environment")=<environment: R_GlobalEnv>
> .. ..- attr(*, "predvars")= language list(speed, dist)
> .. ..- attr(*, "dataClasses")= Named chr [1:2] "numeric" "numeric"
> .. .. ..- attr(*, "names")= chr [1:2] "speed" "dist"
> $ model :'data.frame': 50 obs. of 2 variables:
> ..$ speed: num [1:50] 4 4 7 7 8 9 10 10 10 11 ...
> ..$ dist : num [1:50] 2 10 4 22 16 10 18 26 34 17 ...
> ..- attr(*, "terms")=Classes 'terms', 'formula' language speed ~ dist
> .. .. ..- attr(*, "variables")= language list(speed, dist)
> .. .. ..- attr(*, "factors")= int [1:2, 1] 0 1
> .. .. .. ..- attr(*, "dimnames")=List of 2
> .. .. .. .. ..$ : chr [1:2] "speed" "dist"
> .. .. .. .. ..$ : chr "dist"
> .. .. ..- attr(*, "term.labels")= chr "dist"
> .. .. ..- attr(*, "order")= int 1
> .. .. ..- attr(*, "intercept")= int 1
> .. .. ..- attr(*, "response")= int 1
> .. .. ..- attr(*, ".Environment")=<environment: R_GlobalEnv>
> .. .. ..- attr(*, "predvars")= language list(speed, dist)
> .. .. ..- attr(*, "dataClasses")= Named chr [1:2] "numeric" "numeric"
> .. .. .. ..- attr(*, "names")= chr [1:2] "speed" "dist"
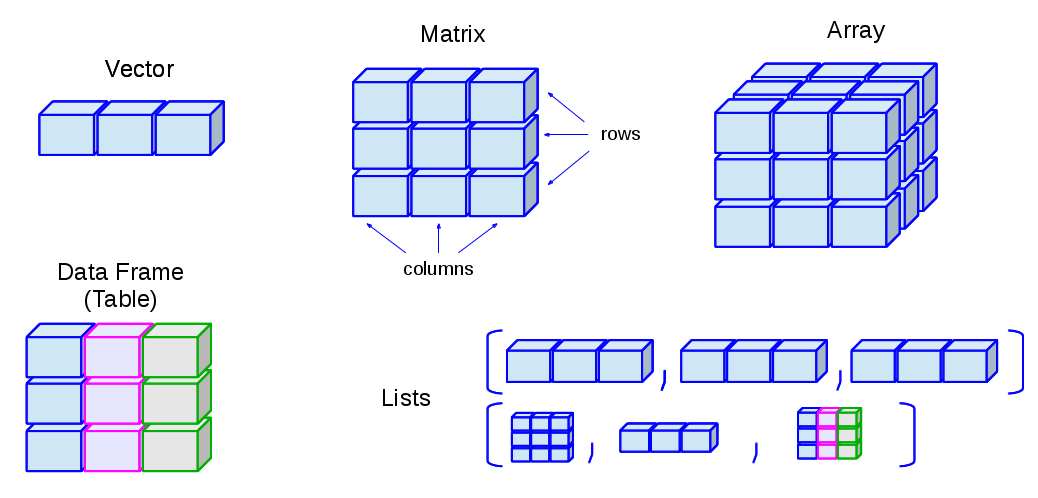
> - attr(*, "class")= chr "lm"Data Structures in R Overview
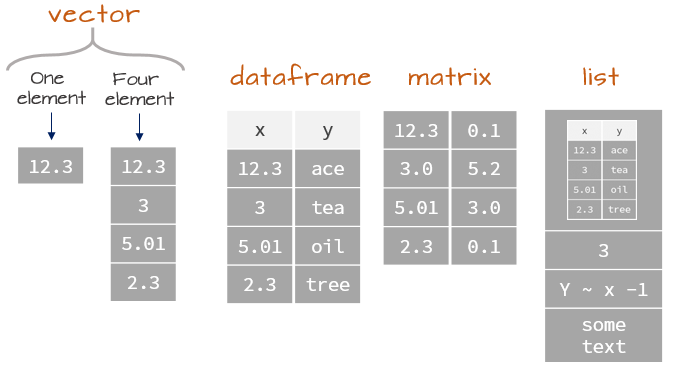
Data Structures in R Overview
Lab
Matrices and Lists
- Write code to create the following matrix:
> [,1] [,2] [,3]
> [1,] "A" "B" "C"
> [2,] "D" "E" "F"- Write a line of code to extract the second column. Ensure the output is still a matrix.
> [,1]
> [1,] "B"
> [2,] "E"Complete the following sentence: “Lists are to vectors, what data frames are to…”
Create a list that contains 3 elements:
ten_numbers(integers between 1 and 10)my_name(your name as a character)booleans(vector ofTRUEandFALSEalternating three times)
Answers
1. Write code to create the following matrix:
> [,1] [,2] [,3]
> [1,] "A" "B" "C"
> [2,] "D" "E" "F"Answers
3. Complete the following sentence: “Lists are to vectors, what data frames are to…Matrices!1”
4. Create a list that contains 3 elements: So many ways to do this! Here’s one example.